Data Exploration#
(15 minutes)

Prerequisites: an EarthData Login
Learning Objectives
Experience a variety of SnowEx data across different spatial and temporal domains
Preview code snippets to help you extract and prep data
Get a first taste of the database!
PART I: Spatial Data#
Let’s take a look at data collected during the Intense Observation Periods (IOP) campaigns via EarthAcess data downloads.
I: 2017/2020 - Grand Mesa & Senator Beck, Colorado#
Objective - Visualize SWE from Grand Mesa field campaigns.
Data description - SnowEx 2017 & 2020 IOP derived SWE from snow pits.
Datasets -
SnowEx17 Community Snow Pit Measurements, Version 1 (Grand Mesa + Senator Beck, CO)
SnowEx20 Grand Mesa Intensive Observation Period Snow Pit Measurements, Version 1 (Grand Mesa, CO)
We will begin by importing python packages we will need to explore data.
# standard imports
import pandas as pd
import pyproj
import datetime
# plotting imports
import matplotlib
import matplotlib.pyplot as plt
# from matplotlib import gridspec # do I use?
import matplotlib.dates as mdates
# import matplotlib.patches as patches # do I use?
import seaborn as sns
# mapping imports
import folium
from pyproj import Transformer
import branca.colormap as cm
from folium import IFrame
from folium.plugins import MeasureControl
# Earthaccess imports
import earthaccess
Now we continue by accessing data with our EarthData accounts from NSIDC.
Download data from NASA’s NSIDC DAAC#
# connect to Earthaccess
auth = earthaccess.login()
From EarthData we can glean information about the files we are interested in. For example, we learn that the SnowEx 2017 snow pit data are a multi-file granule, which will require one extra step to download.
# pull SnowEx 2017 snow pit data - Summary_SWE file
results_gm17 =earthaccess.search_data(
short_name='SNEX17_SNOWPITS',
granule_name = 'SnowEx17_SnowPits_GM')
# some extra code to filter a multi-file granule (2017 IOP granules: 1). SnowEx17_SnowPits_GM, 2). SnowEx17_SnowPits_SBB
filtered_urls = []
for granule in results_gm17:
links = granule.data_links()
for link in links:
if link == 'https://n5eil01u.ecs.nsidc.org/DP1/SNOWEX/SNEX17_SnowPits.001/2017.02.06/SnowEx17_SnowPits_GM_201702_v01.csv':
filtered_urls.append(link) # Knowing the name of the file I wanted helped here
earthaccess.download(filtered_urls,
local_path='./Downloads')
['Downloads/SnowEx17_SnowPits_GM_201702_v01.csv']
# pull SnowEx 2020 snow pit data - Summary_SWE file
results_gm20 =earthaccess.search_data(
short_name='SNEX20_GM_SP',
granule_name = 'SnowEx20_SnowPits_GMIOP_Summary_SWE_2020_v01.csv')
downloaded_file_gm20 = earthaccess.download(
results_gm20,
local_path='./Downloads',
)
Note
The downside to this method is you have to know the specific filename you are interested in. That can require more time to familiarize yourself with the dataset. A great resource is to read the User Guide provided for most SnowEx data sets and accessed from the NSIDC data landing page. The User Guides provide information about filenames.
Look over to the left in your files and see that we added a ./Downloads folder to store the EarthData download.
Next, let’s use Python’s pandas package to open the CSV file that contains Summary SWE data.
# open summary SWE files from our ./Downloads folder
s17 = pd.read_csv('./Downloads/SnowEx17_SnowPits_GM_201702_v01.csv')
s20 = pd.read_csv('./Downloads/SnowEx20_SnowPits_GMIOP_Summary_SWE_2020_v01.csv')
print(s17.info())
s17.describe()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 264 entries, 0 to 263
Data columns (total 13 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 #Location 264 non-null object
1 Site 264 non-null object
2 PitID 264 non-null object
3 Date/Time (MST) 264 non-null object
4 UTM13 Easting [m] 264 non-null int64
5 UTM13 Northing [m] 264 non-null int64
6 Density A Mean [kg/m^3] 264 non-null int64
7 Density B Mean [kg/m^3] 264 non-null int64
8 Density Mean [kg/m^3] 264 non-null int64
9 SWE A [mm] 264 non-null int64
10 SWE B [mm] 264 non-null int64
11 SWE [mm] 264 non-null int64
12 Snow Depth [cm] 264 non-null int64
dtypes: int64(9), object(4)
memory usage: 26.9+ KB
None
| UTM13 Easting [m] | UTM13 Northing [m] | Density A Mean [kg/m^3] | Density B Mean [kg/m^3] | Density Mean [kg/m^3] | SWE A [mm] | SWE B [mm] | SWE [mm] | Snow Depth [cm] | |
|---|---|---|---|---|---|---|---|---|---|
| count | 264.000000 | 2.640000e+02 | 264.000000 | 264.000000 | 264.000000 | 264.000000 | 264.000000 | 264.000000 | 264.000000 |
| mean | 239602.007576 | 4.306328e+06 | 322.227273 | 322.840909 | 322.515152 | 472.484848 | 473.253788 | 472.901515 | 145.856061 |
| std | 12054.013830 | 4.553288e+04 | 17.479849 | 17.394475 | 17.289081 | 123.046437 | 122.673637 | 122.774817 | 34.450412 |
| min | 221314.000000 | 4.196923e+06 | 254.000000 | 263.000000 | 258.000000 | 114.000000 | 118.000000 | 116.000000 | 45.000000 |
| 25% | 232181.000000 | 4.322908e+06 | 313.000000 | 312.000000 | 312.000000 | 399.500000 | 399.000000 | 400.000000 | 126.000000 |
| 50% | 236112.500000 | 4.324478e+06 | 321.000000 | 322.000000 | 321.000000 | 476.000000 | 477.000000 | 477.000000 | 148.500000 |
| 75% | 249477.500000 | 4.326407e+06 | 332.000000 | 334.250000 | 334.000000 | 559.000000 | 563.250000 | 560.000000 | 169.000000 |
| max | 261866.000000 | 4.332499e+06 | 371.000000 | 371.000000 | 371.000000 | 952.000000 | 952.000000 | 952.000000 | 257.000000 |
# some dataframe clean up
s17 = s17[s17['#Location'] == 'GM-CO'] # keep the GM points only for now
s20 = s20[s20['Easting (m)'] < 750000] # removes point at GM lodge (n=1)
# Convert UTM to Lat/Lon
transformer = pyproj.Transformer.from_crs("EPSG:32613", "EPSG:4326") # UTM Zone 13N
s17['latitude'], s17['longitude'] = zip(*s17.apply(lambda row: transformer.transform(row['UTM13 Easting [m]'], row['UTM13 Northing [m]']), axis=1))
transformer = pyproj.Transformer.from_crs("EPSG:32612", "EPSG:4326") # UTM Zone 12N
s20['latitude'], s20['longitude'] = zip(*s20.apply(lambda row: transformer.transform(row['Easting (m)'], row['Northing (m)']), axis=1))
Now that we have added Latitude and Longitude columns, let’s look at the spatial extent of the data.
fig, ax = plt.subplots(figsize=(8,4))
s17.plot.scatter('latitude', 'longitude', color='blueviolet', ax=ax, label='GrandMesa 2017')
s20.plot.scatter('latitude', 'longitude', color='orange', ax=ax, label='GrandMesa 2020')
plt.legend(loc='lower right')
plt.show()
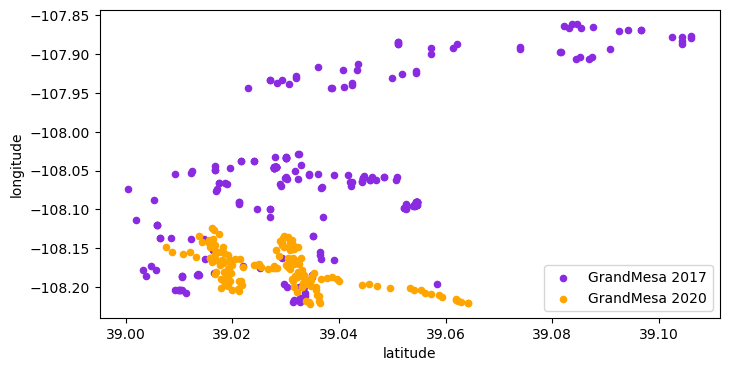
We see that during the 2017 IOP the sampling area covered a greater extent.
Let’s add some context to these data points#
By using an interactive map (Folium) we can move around the Mesa. Here we plot Latitude and Longitude on the y- and x-axes, respectively and add plot SWE as the 3rd dimension with the additional of a colorbar.
# Create a map using lat/lon mean
m = folium.Map(width=500, height=400,location=[s17['latitude'].mean(), s17['longitude'].mean()], zoom_start=10)
m
I used the dataframe’s lat/lon mean to center my map. I choose the 2017 dataframe because of the greater spatial extent we discovered above.
Let’s now modify the map imagery
# Add aerial imagery (ESRI World Imagery)
folium.TileLayer(
tiles='https://server.arcgisonline.com/ArcGIS/rest/services/World_Imagery/MapServer/tile/{z}/{y}/{x}',
attr='Esri',
name='Esri Satellite',
overlay=False,
control=True,
opacity=0.9
).add_to(m)
m
Oh goody, I can see the Mesa! Did you know this is the largest flat-topped mountain in the world? See for yourself right here.
Moving on, let’s add the SWE values to the map!
Map + Imagery + Data#
# Create a Folium map
m = folium.Map(width=1000, height=600, location=[s17['latitude'].mean(), s17['longitude'].mean()], zoom_start=10)
# Add aerial imagery (ESRI World Imagery)
folium.TileLayer(
tiles='https://server.arcgisonline.com/ArcGIS/rest/services/World_Imagery/MapServer/tile/{z}/{y}/{x}',
attr='Esri',
name='Esri Satellite',
overlay=False,
control=True,
opacity=0.9
).add_to(m)
# Create a colormap
colormap1 = cm.linear.Purples_09.to_step(12).scale(50,800)
colormap2 = cm.linear.YlOrBr_04.to_step(12).scale(50,800)
# Plot the scatter plot for s17
for index, row in s17.iterrows():
folium.CircleMarker(location=[row['latitude'], row['longitude']], radius=3, fill=True, fill_opacity=1, color=colormap1(row['SWE [mm]'])).add_to(m)
# Plot the scatter plot for s20
for index, row in s20.iterrows():
folium.CircleMarker(location=[row['latitude'], row['longitude']], radius=3, fill=True, fill_opacity=1, color=colormap2(row['SWE Mean (mm)'])).add_to(m)
# Add colorbar legend to the map
colormap1.add_to(m)
colormap1.caption = '2017 SWE (mm)'
colormap1.stroke_color = '#ffffff' # White color for the text buffer
colormap2.add_to(m)
colormap2.caption = '2020 SWE (mm)'
colormap2.stroke_color = '#ffffff' # White color for the text buffer
# Define custom CSS for the colorbar legend
custom_css = """
<style>
.legend {
font-size: 16px !important;
height: 80px !important; /* Adjust the height of the colorbar */
}
.legend .caption {
font-size: 18px !important;
font-weight: bold;
text-shadow: 0 0 3px #ffffff;
}
.legend .min, .legend .max {
font-size: 16px !important;
text-shadow: 0 0 3px #ffffff;
}
</style>
"""
m.add_child(MeasureControl(position='bottomleft', primary_length_unit='meters', secondary_length_unit='miles', primary_area_unit='sqmeters', secondary_area_unit='acres'))
# Add the custom CSS to the map
m.get_root().html.add_child(folium.Element(custom_css))
# Save the map as an HTML file or display it in Jupyter notebook
# m.save(path_out.joinpath('GMIOP_20_swe_map.html'))
# Display the map
m
Note
The Grand Mesa plateau splits the 12 & 13 UTM zone. That means the western portion of the Mesa is Zone 12 while the eastern portion is Zone 13. Due to the sampling design, the SnowEx 2017 data were collected as Zone 13 (majority of points were east of the divide) and SnowEx 2020 data were collected as Zone 12 (majority of points focused on the western portion of the plateau).
What variability is there during the different sampling years?#
# open again to reset (recall, we filtered out Senator Beck data before, let's keep that this time!)
s17 = pd.read_csv('./Downloads/SnowEx17_SnowPits_GM_201702_v01.csv')
s20 = pd.read_csv('./Downloads/SnowEx20_SnowPits_GMIOP_Summary_SWE_2020_v01.csv')
# subdataframes for GM and SB for 2017
s17_gm = s17[s17['#Location'] == 'GM-CO']
s17_sb = s17[s17['#Location'] == 'SBB-CO']
fig, axs = plt.subplots(1, 2, figsize=(12, 5))
# ~~~~ Plot 1: Scatter plot of density and SWE for S17 and S20
# Count the number of observations
n_s17_sb = s17_sb['SWE [mm]'].count()
n_s17_gm = s17_gm['SWE [mm]'].count()
n_s20 = s20['SWE Mean (mm)'].count()
# Plot data for 2017 SBB
sns.scatterplot(data=s17_sb, x='Density Mean [kg/m^3]', y='Snow Depth [cm]', s=60, label=f'2017 SBB-CO (n={n_s17_sb})', marker='^', ax=axs[0])
# Plot data for 2017 GM
sns.scatterplot(data=s17_gm, x='Density Mean [kg/m^3]', y='Snow Depth [cm]', s=60, label=f'2017 GM-CO (n={n_s17_gm})', marker='^', ax=axs[0])
# Plot data for 2020 GM
sns.scatterplot(data=s20, x='Density Mean (kg/m^3)', y='Snow Depth (cm)', s=40, label=f'2020 GM-CO (n={n_s20})', ax=axs[0])
# Adding labels and legends
axs[0].set_xlabel('Snow Density (kg/m$^3$)')
axs[0].set_ylabel('Snow Depth (cm)')
axs[0].legend()
axs[0].set_title('S17 and S20 Snow Pit Bulk Density and Depth')
axs[0].grid(True)
# ~~~~ Plot 2: PDF for S17 (Grand Mesa + Senator Beck) and S20 (Grand Mesa 2020)
# Plot the PDF for 2017 SBB
sns.kdeplot(s17_sb['SWE [mm]'], label='2017 SBB-CO', linewidth=5, ax=axs[1])
# Plot the PDF for 2017 GM
sns.kdeplot(s17_gm['SWE [mm]'], label='2017 GM-CO', linewidth=5, ax=axs[1])
# Plot the PDF for 2020 GM
sns.kdeplot(s20['SWE Mean (mm)'], label='2020 GM-CO', linewidth=5, ax=axs[1])
# Add title and labels
axs[1].set_title('S17 and S20 Snow Pit SWE')
axs[1].set_xlabel('SWE (mm)')
axs[1].set_ylabel('PDF')
axs[1].legend()
# Save figure if needed
# plt.savefig(path_fig.joinpath('S17_n_S20_GMIOP_pdf.jpg'), bbox_inches='tight', pad_inches=0.1)
# Show the plot
plt.tight_layout()
plt.show()
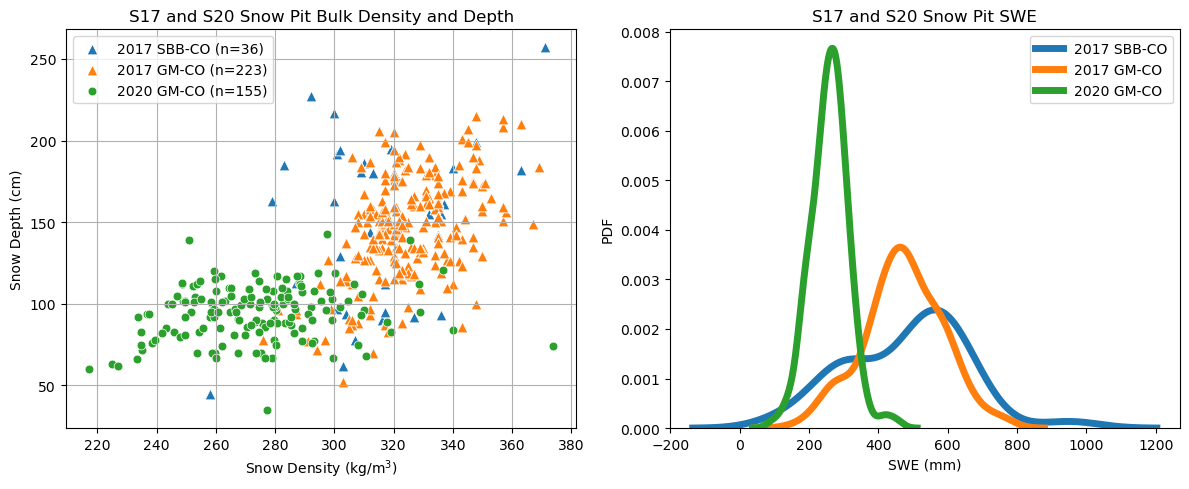
There is so much more we could dig into. Just ask HP, he’ll say you could spend a lifetime studying snow on Grand Mesa :)
Alas, let us continue our spatial exploration but move it up to Alaska.
II: 2023 Arctic Tundra and Boreal Forest, Alaska#
Objectives:
Compare snow depth, density and SWE data from the Adirondack snow sampler (n=84) and snow pits (n=170)
Explore data within the
snowexsqldatabase Data description - SnowEx 2023 March 2023 IOP Snow Water Equivalent and Snow Pit Measurements\
Datasets -
SnowEx23 Snow Water Equivalent, Version 1 (2-days of Adirondack snow sampler measurements North Slope (tundra) and Fairbanks (boreal forest))
SnowEx23 Snow Pit Measurements, Version 1 (in prep., available on the
snowexsqldatabase)
# Adirondack SWE download
results_swe = earthaccess.search_data(
short_name='SNEX23_SWE')
downloaded_files_swe = earthaccess.download(
results_swe,
local_path='./Downloads',
)
# datasets from Alaska contain more header rows. skipping 42 rows for now and dropping the numbered row under the columns names
s23 = pd.read_csv('./Downloads/SNEX23_SWE_Mar23IOP_AK_20230313_20230316_v01.0.csv', header=42, na_values=-9999)
s23 = s23.iloc[1:] # selects the row below the header number cols(1, 2, 3, ... etc.)
s23 = s23.reset_index(drop=True)
s23['County'] = s23['County'].replace({'Fairbanks': 'FLCF \n(Boreal Forest)', 'North Slope': 'UKT \n(Arctic Tundra)'})
s23.head()
| State | County | Site ID | Latitude | Longitude | Elevation | Date | Depth1 | Depth2 | Depth3 | Mass1 | Mass2 | Mass3 | Density1 | Density2 | Density3 | SWE1 | SWE2 | SWE3 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Alaska | UKT \n(Arctic Tundra) | UKE1 | 68.611087 | -149.493961 | 963 | 13-Mar-23 | 34 | 32.0 | 32.0 | 198 | 190.0 | 209.0 | 163 | 166.0 | 183.0 | 55 | 53.0 | 59.0 |
| 1 | Alaska | UKT \n(Arctic Tundra) | UKE1.5 | 68.611280 | -149.481889 | 942 | 13-Mar-23 | 72 | 76.0 | 74.0 | 645 | 663.0 | 584.0 | 251 | 244.0 | 303.0 | 181 | 186.0 | 164.0 |
| 2 | Alaska | UKT \n(Arctic Tundra) | UKE2 | 68.611497 | -149.470469 | 935 | 13-Mar-23 | 97 | 102.0 | 101.0 | 839 | 891.0 | 917.0 | 242 | 245.0 | 254.0 | 235 | 250.0 | 257.0 |
| 3 | Alaska | UKT \n(Arctic Tundra) | UKE2.5 | 68.611411 | -149.459072 | 951 | 13-Mar-23 | 32 | 20.0 | 12.0 | 188 | 91.0 | 85.0 | 165 | 127.0 | 198.0 | 53 | 25.0 | 24.0 |
| 4 | Alaska | UKT \n(Arctic Tundra) | UKE3 | 68.611550 | -149.447256 | 938 | 13-Mar-23 | 68 | 68.0 | 68.0 | 502 | 484.0 | 470.0 | 207 | 201.0 | 197.0 | 141 | 137.0 | 134.0 |
At each coordinate pair, the Adirondack snow sampler (i.e a large SWE tube, cross-sectional area A = 35.7 cm2) is used to collect 3 samples each of depth (cm) and mass (g). Bulk snow density and SWE are then computed afterwards using the equations from Stuefer et al., 2020. Here is the device being used on the North Slope in Alaska.

Okay, back to some data analysis.
# CREATE DEPTH, DENSITY, and SWE Boxplots with the SWE Adirondack snow sampler
# Combine the SWE, Depth, and Density columns for plotting (3 samples/location)
df_melted = pd.melt(s23, id_vars=['State', 'County'], value_vars=['SWE1', 'SWE2', 'SWE3', 'Depth1', 'Depth2', 'Depth3', 'Density1', 'Density2', 'Density3'],
var_name='Measurement', value_name='Value')
# Create sub-dataframes for SWE, Depth, and Density
swe_df = df_melted[df_melted['Measurement'].str.contains('SWE')]
depth_df = df_melted[df_melted['Measurement'].str.contains('Depth')]
density_df = df_melted[df_melted['Measurement'].str.contains('Density')]
# Initialize the figure and axes
fig, axs = plt.subplots(1, 3, figsize=(14, 6))
# Define the custom order and labels
custom_order = ['FLCF \n(Boreal Forest)', 'UKT \n(Arctic Tundra)']
# Plot the SWE boxplot
sns.boxplot(data=swe_df, x='County', y='Value', ax=axs[0], color='gray', order=custom_order)
axs[0].set_title('SWE')
axs[0].set_ylabel('SWE (mm)')
# Plot the Depth boxplot
sns.boxplot(data=depth_df, x='County', y='Value', ax=axs[1], color='gray', order=custom_order)
axs[1].set_title('Depth')
axs[1].set_ylabel('Depth (cm)')
# Plot the Density boxplot
sns.boxplot(data=density_df, x='County', y='Value', ax=axs[2], color='gray', order=custom_order)
axs[2].set_title('Density')
axs[2].set_ylabel('Density (kg/m$^3$)')
# Adjust the layout
plt.tight_layout()
plt.show()
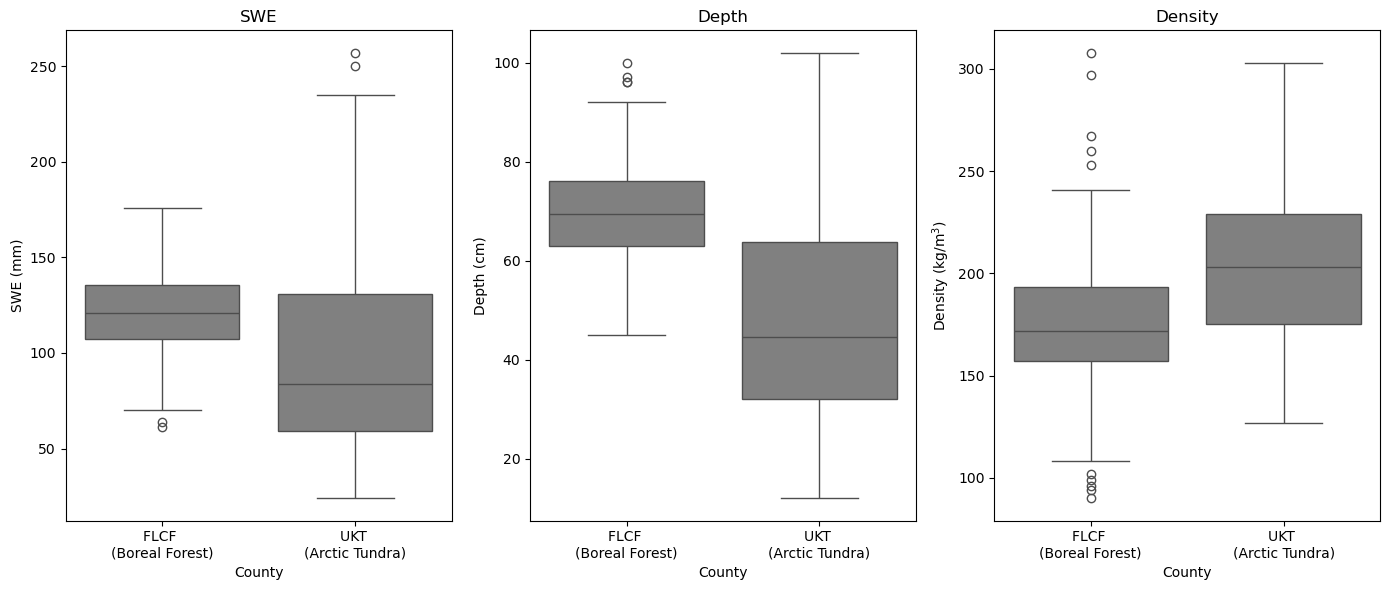
We can compare our Adirondack snow sampler results to the snow pit measurement. The boxplots below were generated during the review and data submission preparation work for the March 2023 snow pits. Notice, the snow pit trends match those of the snow sampler tube. The median SWE is higher in the boreal forest as a result of the deeper snow. Overall, the tundra sites have greater density.
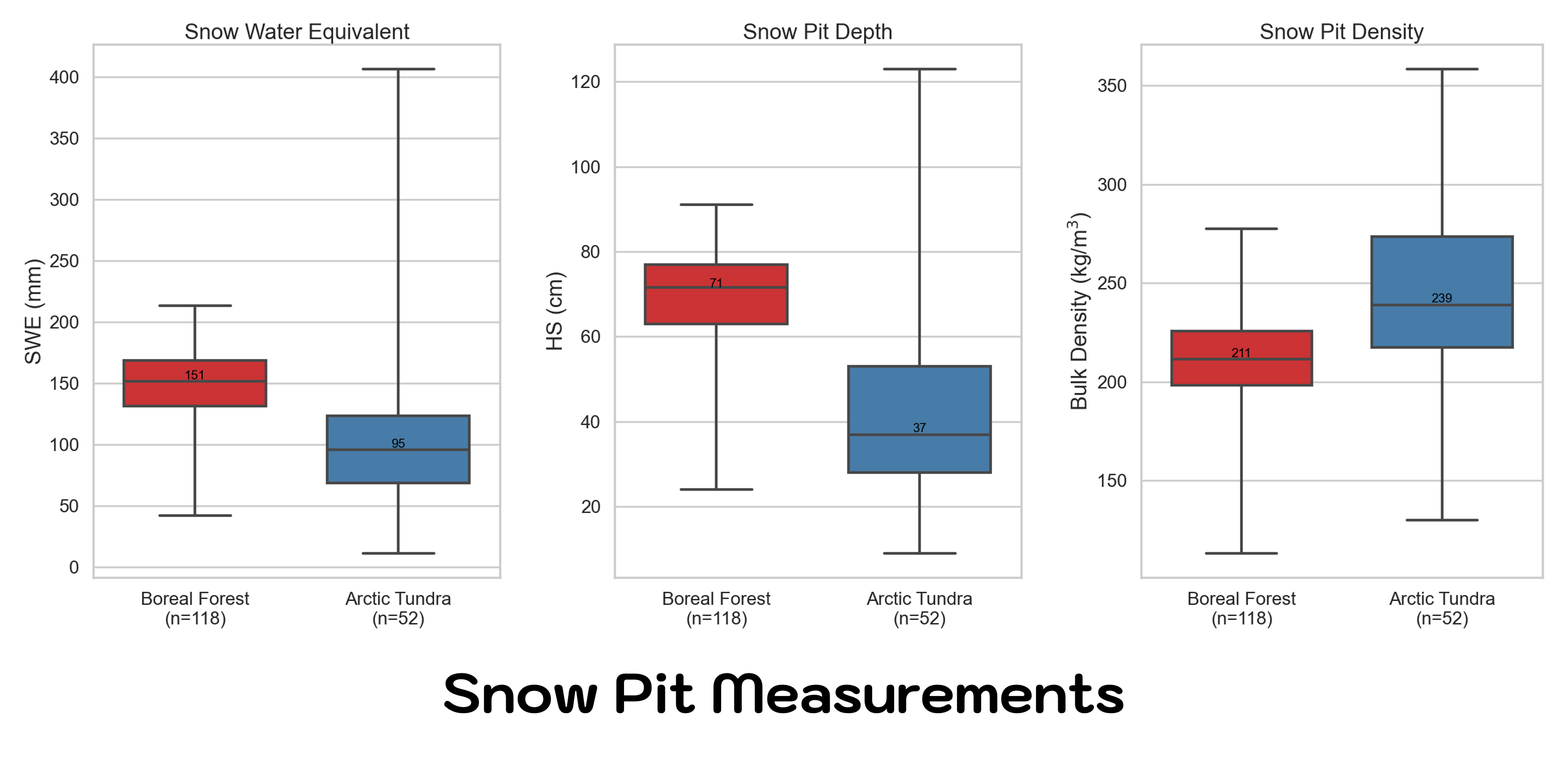
What are other ways to look at the data? Recall that the Boreal Forest has a land cover classification enbedded in the plot name. Why not look at SWE, depth, and density by land cover type? Recall, the Land Classification information is embedded in the pitID name (e.g. WB241 the ‘W’ stands for wetland, or EA221 the ‘E’ stands for Evergreen).
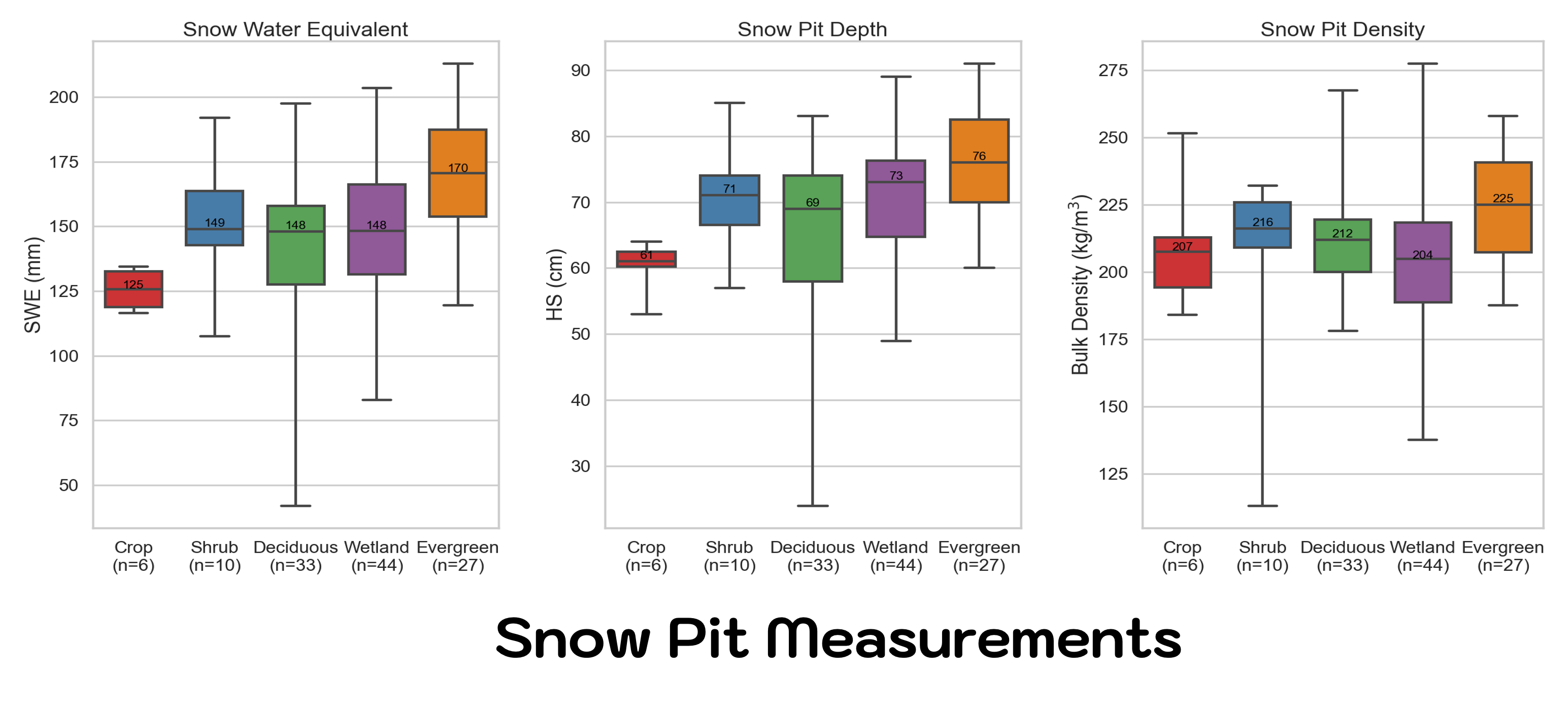
Does this mean that all Evergreen sites have more snow!?
Careful now, there is a lot to consider when interpreting results. This plot alone looks like we have deeper, denser snow in the Evergreen forests, but keep in mind you may not know the other environmental and terrain factors that might cause you to jump to this conclusion. Keep in mind relative location and elevation when looking at data. The Alaska study sites are all relativily flat compared to traditional complex terrain areas, but the majority of our Evergreen sites were sampled in the Caribou-Poker Creek Research Watershed, which is higher in elevation that the sites closer to Fairbanks.
Alaska was the last stop for SnowEx field campaigns, that means the data collection was easy-breezy.
WRONG.
Here are some challenges we encountered daily:
Ground vegetation - where is 0? Often, the bottom of the snowpack varied more than the snow surface!
Tussocks, what are they and why are there so many? (NOTE - tussocks were better quantified during the October sampling periods when snow was low.
Snow Voids - snowpack bottoms were not always flush to the ground surface and could be overestimated. Grassy vegetation mats, downed brush and shrubs, or other vegetation creates air pockets or vegetation gaps near the ground surface. Sometimes, the snow was suspended by 10-25 cm depending on the site.
These data are really exciting to look at because it’s the newest in the SnowEx collection following the Alaska campaign.
Let me introduce the snowexsql database by demonstrating how to run an example query. To work from the database, we are going to add some necessary imports.
Set Up Computing Environment#
#database imports
from snowexsql.db import get_db
from snowexsql.data import PointData, LayerData, ImageData, SiteData
from snowexsql.conversions import query_to_geopandas
The Basics#
How are the data contained?#
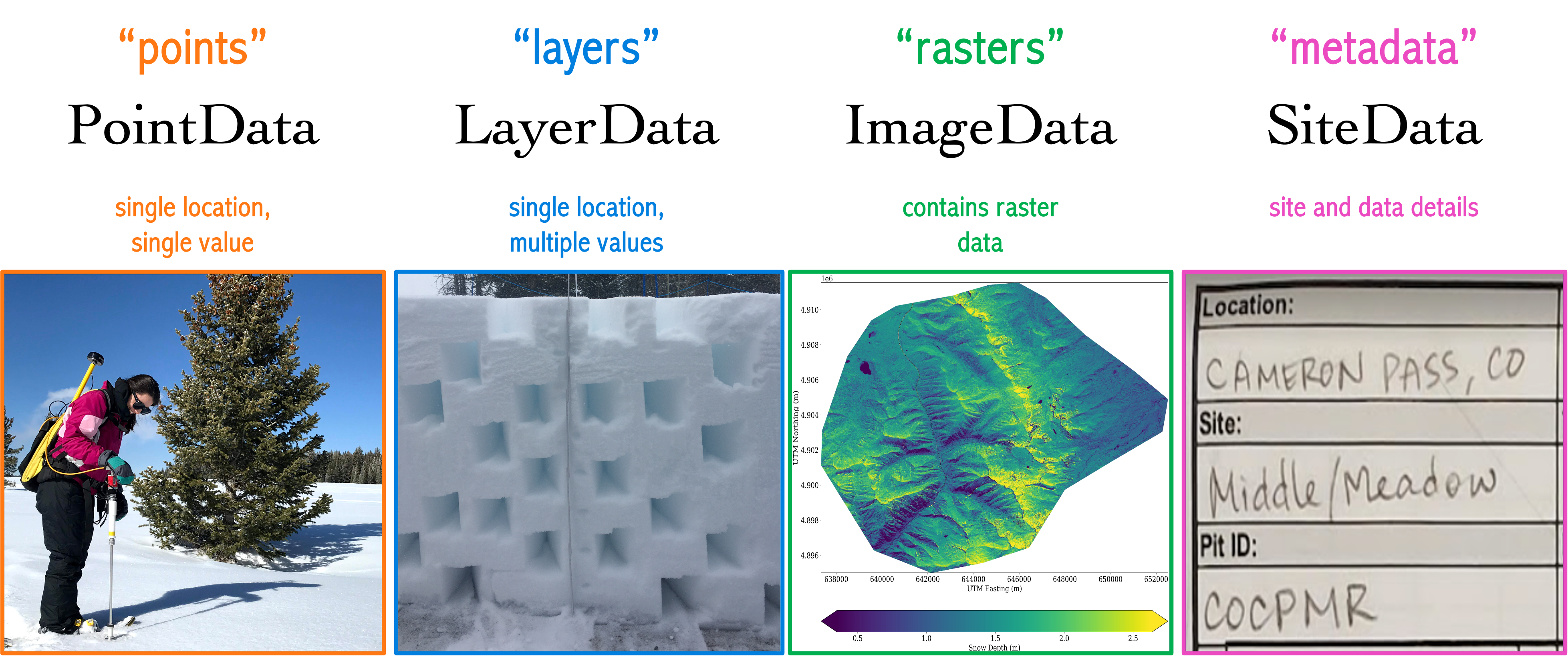
# load the database
db_name = 'snow:hackweek@db.snowexdata.org/snowex'
engine, session = get_db(db_name)
print('SnowEx Database successfully loaded!')
SnowEx Database successfully loaded!
What’s the first thing you might like to do using the database?#
Find overlapping data for data analysis comparison.
Example 1: Let’s find all the pits that overlap with an airborne sensor of interest.#
First, it would be helpful to know which of the airborne sensors are part of the database, right?
# Query the session using .observers() to generate a list
qry = session.query(ImageData.observers)
# Locate all that are distinct
airborne_sensors_list = session.query(ImageData.observers).distinct().all()
print('list of airborne sensors by "observer" name: \n', airborne_sensors_list)
list of airborne sensors by "observer" name:
[('USGS',), ('UAVSAR team, JPL',), ('ASO Inc.',), ('chris larsen',)]
Note: ‘chris larsen’ == airborne lidar data from Alaska
1a). Unsure of the flight date, but know which sensor you’d like to overlap with, here’s how:#
# Airborne sensor from list above
sensor = 'chris larsen' # Name given to Alaska Lidar data, observer Dr. Chris Larsen
# Form a query on the Images table that returns Raster collection dates
qry = session.query(ImageData.date)
# Filter for Alaska Lidar data
qry = qry.filter(ImageData.observers == sensor)
# Grab the unique dates
qry = qry.distinct()
# Execute the query
dates = qry.all()
# Clean up the dates
dates = [d[0] for d in dates]
dlist = [str(d) for d in dates]
dlist = ", ".join(dlist)
print('%s flight dates are: %s' %(sensor, dlist))
# Find all the snow pits done on these days
qry = session.query(SiteData.geom, SiteData.site_id, SiteData.date)
qry = qry.filter(SiteData.date.in_(dates))
# Return a geopandas df
df = query_to_geopandas(qry, engine)
# View the returned dataframe!
print(df.head())
print(f'{len(df.index)} records returned')
# Close your session to avoid hanging transactions
session.close()
chris larsen flight dates are: 2022-10-24, 2023-03-11
geom site_id date
0 POINT (406069.144 7611197.807) UKT 2023-03-11
1 POINT (438495.836 7775803.918) ACP 2023-03-11
2 POINT (406060.880 7611607.651) UKT 2023-03-11
3 POINT (438654.824 7776090.881) ACP 2023-03-11
4 POINT (405487.123 7613118.982) UKT 2023-03-11
68 records returned
1b).Want to select an exact flight date match? Here’s how:#
# Pick a day from the list of dates
dt = dates[1] # selecting the "2nd" date in the list, 203-03-11 during the March 2023 field campaign
# Find all the snow pits done on these days
qry = session.query(SiteData.geom, SiteData.site_id, SiteData.date)
qry = qry.filter(SiteData.date == dt)
# Return a geopandas df
df_exact = query_to_geopandas(qry, engine)
print('%s pits overlap with %s on %s' %(len(df_exact), sensor, dt))
# View snows pits that align with first UAVSAR date
df_exact.head()
68 pits overlap with chris larsen on 2023-03-11
| geom | site_id | date | |
|---|---|---|---|
| 0 | POINT (406069.144 7611197.807) | UKT | 2023-03-11 |
| 1 | POINT (438495.836 7775803.918) | ACP | 2023-03-11 |
| 2 | POINT (406060.880 7611607.651) | UKT | 2023-03-11 |
| 3 | POINT (438654.824 7776090.881) | ACP | 2023-03-11 |
| 4 | POINT (405487.123 7613118.982) | UKT | 2023-03-11 |
1c). Want to further refine your search to select data from a specific Alaska study site? Here’s how:#
# Refine your search to Farmers Loop and Creamers Field
site = 'FLCF'
qry = qry.filter(SiteData.site_id == site)
# Return a geopandas df
df_exact = query_to_geopandas(qry, engine)
print('%s pits overlap with %s at %s' %(len(df_exact), sensor, site))
# View snows pits that align with first UAVSAR date
df_exact.head()
16 pits overlap with chris larsen at FLCF
| geom | site_id | date | |
|---|---|---|---|
| 0 | POINT (467094.944 7194364.336) | FLCF | 2023-03-11 |
| 1 | POINT (467087.000 7194547.208) | FLCF | 2023-03-11 |
| 2 | POINT (467628.759 7194826.632) | FLCF | 2023-03-11 |
| 3 | POINT (467182.879 7194432.474) | FLCF | 2023-03-11 |
| 4 | POINT (467094.944 7194364.336) | FLCF | 2023-03-11 |
1d). Have a known date that you wish to select data for, here’s how:#
# Find all the data that was collected on 3-11-2023 (i.e. the same day we selected airborne lidar data for)
dt = datetime.date(2023, 3, 11)
#--------------- Point Data -----------------------------------
# Grab all Point data instruments from our date
point_instruments = session.query(PointData.instrument).filter(PointData.date == dt).distinct().all()
point_type = session.query(PointData.type).filter(PointData.date == dt).distinct().all()
# Clean up point data (i.e. remove tuple)
point_instruments = [p[0] for p in point_instruments if p[0] is not None]
point_instruments = ", ".join(point_instruments)
point_type = [p[0] for p in point_type]
point_type = ", ".join(point_type)
print('Point data on %s are: %s, with the following list of parameters: %s' %(str(dt), point_instruments, point_type))
#--------------- Layer Data -----------------------------------
# Grab all Layer data instruments from our date
layer_instruments = session.query(LayerData.instrument).filter(LayerData.date == dt).distinct().all()
layer_type = session.query(LayerData.type).filter(LayerData.date == dt).distinct().all()
# Clean up layer data
layer_instruments = [l[0] for l in layer_instruments if l[0] is not None]
layer_instruments = ", ".join(layer_instruments)
layer_type = [l[0] for l in layer_type]
layer_type = ", ".join(layer_type)
print('\nLayer Data on %s are: %s, with the following list of parameters: %s' %(str(dt), layer_instruments, layer_type))
#--------------- Image Data -----------------------------------
# Grab all Image data instruments from our date
image_instruments = session.query(ImageData.instrument).filter(ImageData.date == dt).distinct().all()
image_type = session.query(ImageData.type).filter(ImageData.date == dt).distinct().all()
# Clean up image data
image_instruments = [i[0] for i in image_instruments]
image_instruments = ", ".join(image_instruments)
image_type = [i[0] for i in image_type if i[0] is not None]
image_type = ", ".join(image_type)
print('\nImage Data on %s are: %s, with the following list of parameters: %s' %(str(dt), image_instruments, image_type))
Point data on 2023-03-11 are: pulseEkko pro 1 GHz GPR, with the following list of parameters: density, depth, snow_void, swe, two_way_travel
Layer Data on 2023-03-11 are: , with the following list of parameters: density, grain_size, grain_type, hand_hardness, lwc_vol, manual_wetness, permittivity, temperature
Image Data on 2023-03-11 are: lidar, with the following list of parameters: depth
# fix this
qry = session.query(LayerData.value).filter(LayerData.type=='grain_type')
qry = qry.filter(LayerData.value=='MFcr')
count = qry.count()
print('The number of Layer Data containing melt-freeze crusts (MFcr) on %s are: %s' %(str(dt), count))
session.close()
The number of Layer Data containing melt-freeze crusts (MFcr) on 2023-03-11 are: 625
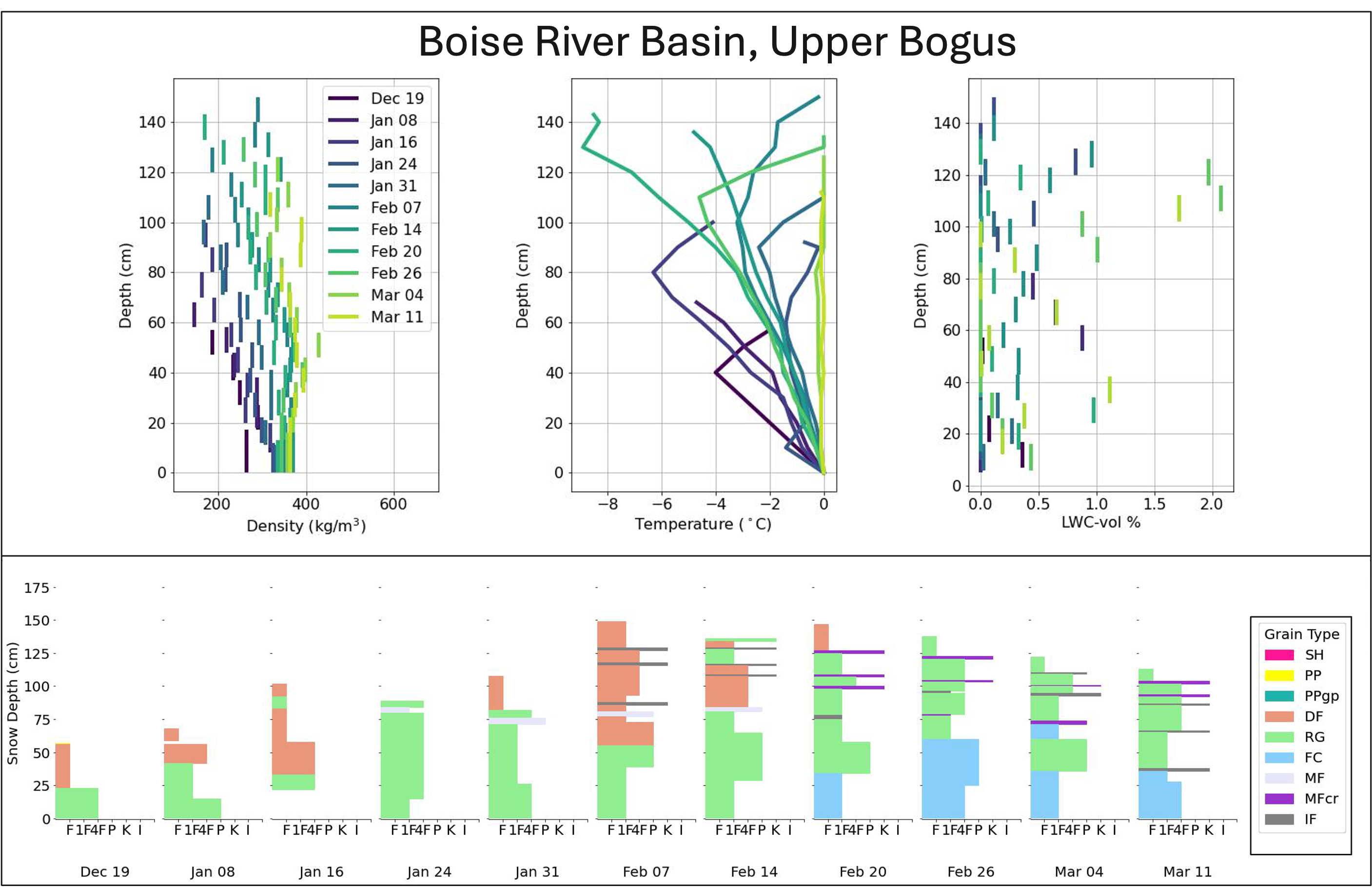
For more useful examples of querying and plotting please visit previous tutorial materials:#
Hackweek 2022 - 03_snowex_database_preview (Examples #2 & #3)
Example #2 - plot snow depth profiles from open Time Series sites.
Example #3 - search for available liquid water content data through an interactive map.
Recap#
You know how to access NSIDC data and interact with 2017 & 2020 Grand Mesa and Senator Beck, Colorado SWE data.
You are aware of some general trends in tundra vs. boreal forest data and can form queries using the
snowexsqldatabase.You have knowledge of what can be harnessed from the 2020 & 2021 Western U.S Time Series campaigns.